Inspired by openair::timeVariation() multiple functions to plot diurnal,
day hour of the week, day of the week and monthly variation. The functions are
optimized for data in long format and the defaults are for the rolf format from
the rOstluft Package.
Usage
gg_timevariation(
data,
dt = "starttime",
y = "value",
group = NULL,
id_cols = grp("site", "parameter", "interval", "unit"),
statistic = c("mean", "median"),
draw_ci = TRUE,
conf_interval = NULL,
B = 1000,
ylab = ggplot2::waiver(),
ylim = c(NA, NA),
legend_title = NULL,
language_code = "de",
geom = ggplot2::geom_line(size = 1),
geom_ci = ggplot2::geom_ribbon(alpha = 0.2)
)
gg_timevariation_wday_hour(
data,
dt = "starttime",
y = "value",
group = NULL,
id_cols = grp("site", "parameter", "interval", "unit"),
statistic = c("mean", "median"),
draw_ci = TRUE,
conf_interval = NULL,
ylab = ggplot2::waiver(),
B = 1000,
ylim = c(NA, NA),
legend_title = NULL,
language_code = "de",
geom = ggplot2::geom_line(size = 1),
geom_ci = ggplot2::geom_ribbon(alpha = 0.2)
)
gg_timevariation_wday(
data,
dt = "starttime",
y = "value",
group = NULL,
id_cols = grp("site", "parameter", "interval", "unit"),
statistic = c("mean", "median"),
draw_ci = TRUE,
conf_interval = NULL,
B = 1000,
ylab = ggplot2::waiver(),
ylim = c(NA, NA),
legend_title = NULL,
language_code = "de",
geom = ggplot2::geom_line(size = 1),
geom_ci = ggplot2::geom_ribbon(alpha = 0.2)
)
gg_timevariation_month(
data,
dt = "starttime",
y = "value",
group = NULL,
id_cols = grp("site", "parameter", "interval", "unit"),
statistic = c("mean", "median"),
draw_ci = TRUE,
conf_interval = NULL,
B = 1000,
ylab = ggplot2::waiver(),
ylim = c(NA, NA),
legend_title = NULL,
language_code = "de",
geom = ggplot2::geom_line(size = 1),
geom_ci = ggplot2::geom_ribbon(alpha = 0.2)
)
gg_timevariation_diurnal(
data,
dt = "starttime",
y = "value",
group = NULL,
id_cols = grp("site", "parameter", "interval", "unit"),
statistic = c("mean", "median"),
draw_ci = TRUE,
conf_interval = NULL,
B = 1000,
ylab = ggplot2::waiver(),
ylim = c(NA, NA),
legend_title = NULL,
language_code = "de",
geom = ggplot2::geom_line(size = 1),
geom_ci = ggplot2::geom_ribbon(alpha = 0.2)
)Arguments
- data
data frame with input data as hourly time series
- dt
a string or symbol for the date time column (default: starttime)
- y
a string or symbol specifying the target column to be summarised (default: value)
- group
column as string to be used to split the input data in multiple groups. Should be a member of
id_colseg. parameter or site (default NULL)- id_cols
A set of columns that uniquely identifies each observation. Use
rOstluft.plot::grp()for quoting. defaultrOstluft.plot::grp(site, parameter, interval, unit)- statistic
Can be
“mean”(default) or“median”. If the statistic is ‘mean’ then the mean line and the 95% confidence interval in the mean are plotted by default.ggplot2::mean_cl_boot()is used to calculated the intervals trough bootstrap simulations without assuming normality. If the statistic is ‘median’ then the median line is plotted together with the 25/75th quantiles are plotted. Users can control the confidence intervals withdraw_ciandconf_interval- draw_ci
if
TRUEdraw confidence interval usingggplot2::mean_cl_boot()for"mean"andggplot2::median_hilow()for"median".- conf_interval
for
"mean"(ggplot2::mean_cl_boot()) specifies the confidence level (0-1) for interval estimation of the population mean. For"median"(ggplot2::median_hilow()) conf_interval is the coverage probability the outer quantiles should target. When the default, 0.5, is used, the lower and upper quantiles computed are 0.25 and 0.75- B
number of bootstrap resamples for
ggplot2::mean_cl_boot()- ylab
provide a custom y plot label
- ylim
limits for y scale see
ggplot2::scale_y_continuous()for more infos.- legend_title
provide a legend title
- language_code
ISO country code for the language used as weekdays and months labels (default: "de")
- geom
geom used for rendering default
ggplot2::geom_line()- geom_ci
geom used for rendering confidence interval. Must support ymin/ymax mapping. default
ggplot2::geom_ribbon()
Value
a ggplot2::ggplot() object or in case of gg_timevariaton() a patchwork::patchwork object
Examples
library(ggplot2)
fn <- rOstluft.data::f("Zch_Stampfenbachstrasse_2010-2014.csv")
data <-
rOstluft::read_airmo_csv(fn) %>%
rOstluft::pluck_parameter("NOx", "NO", "NO2") %>%
rOstluft::resample(new_interval = "h1")
# monthly variation of data
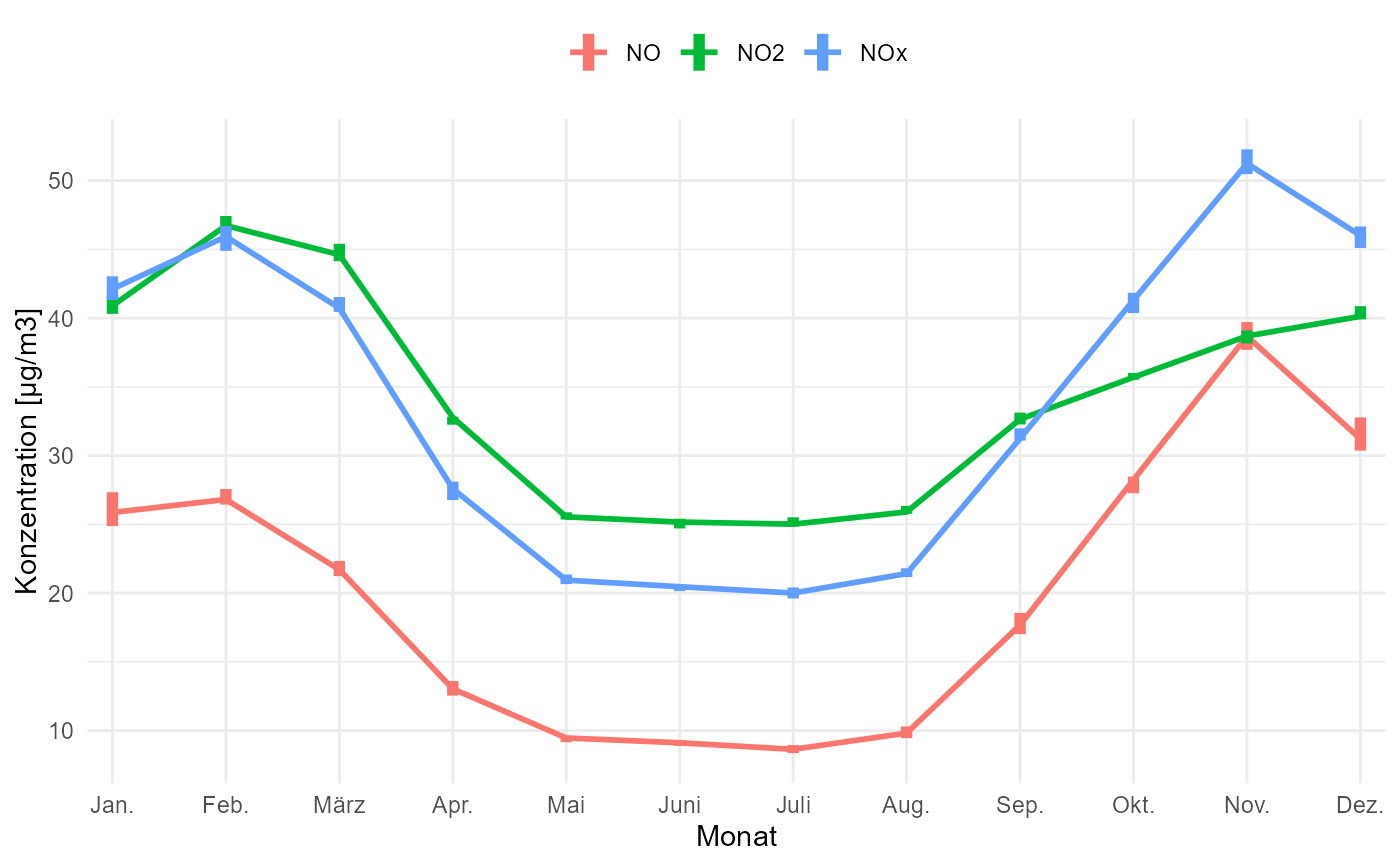
gg_timevariation_month(data, group = "parameter", ylab = "Konzentration [µg/m3]")
#> plotting with statistic mean and confidence interval of 0.95
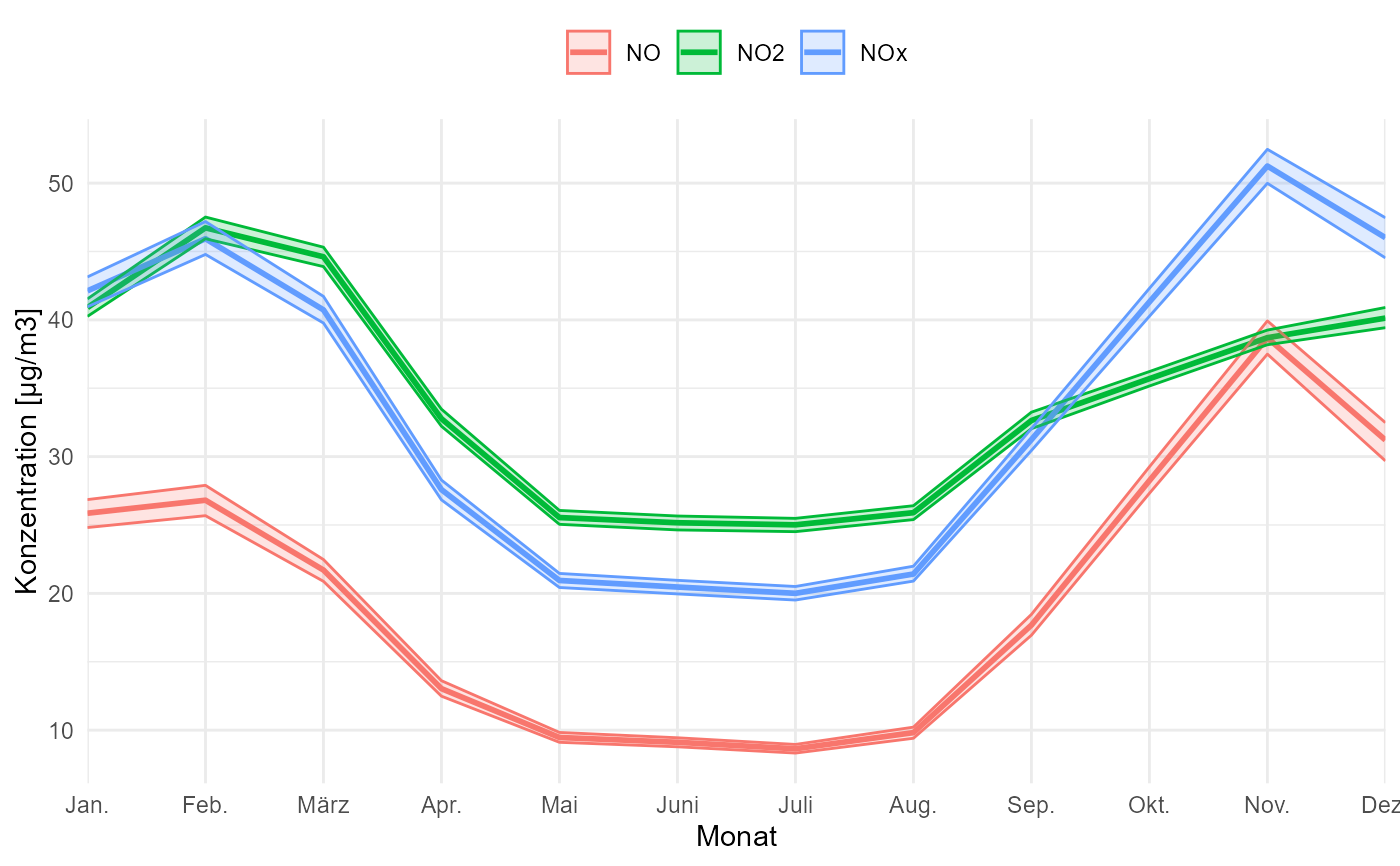 # don't draw a confidence interval
gg_timevariation_month(data, group = "parameter", draw_ci = FALSE, ylab = "Konzentration [µg/m3]")
# don't draw a confidence interval
gg_timevariation_month(data, group = "parameter", draw_ci = FALSE, ylab = "Konzentration [µg/m3]")
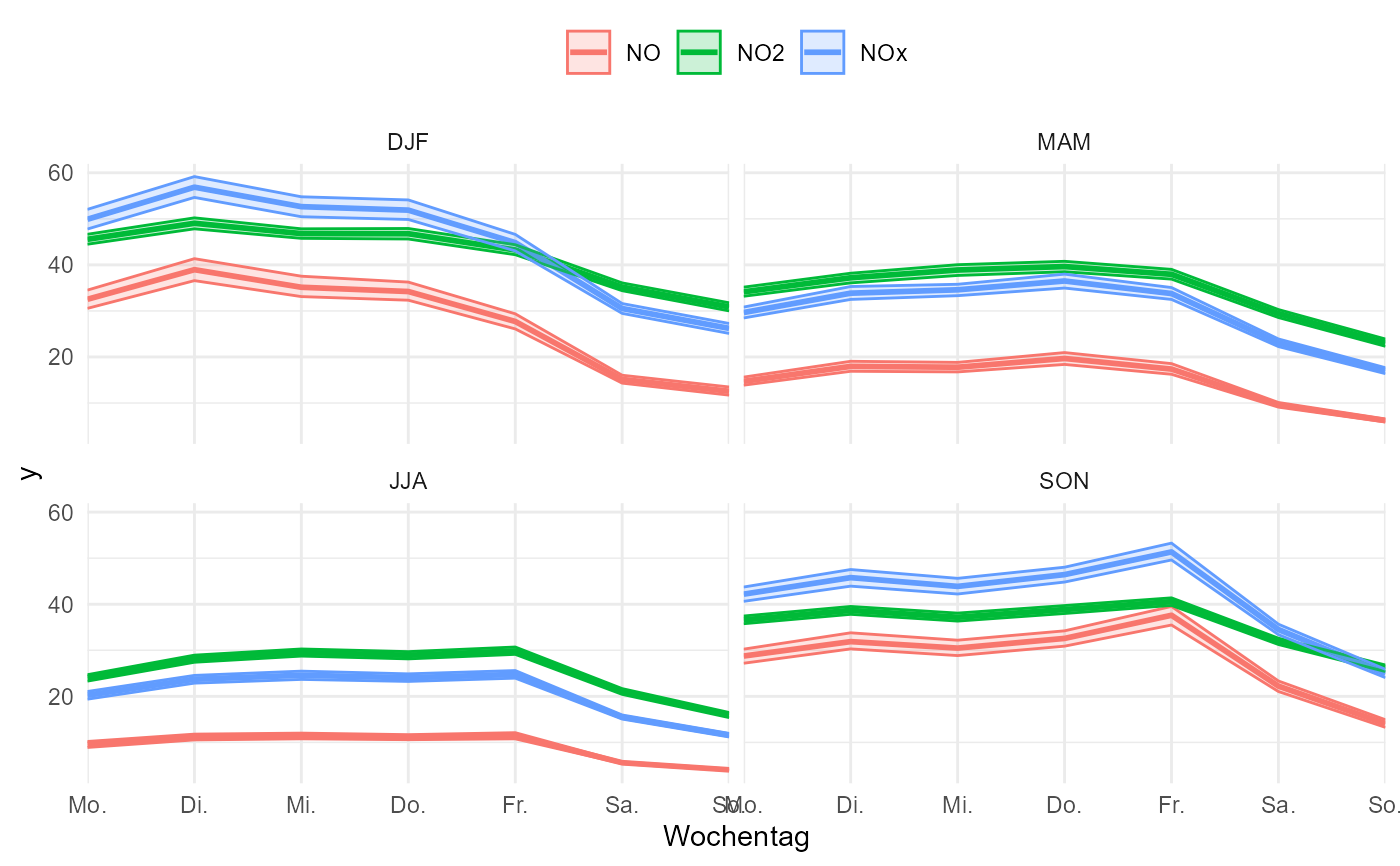
 # for faceting the variable must be included in the id_cols
gg_timevariation_wday(
data,
group = "parameter",
id_cols = grp(site, parameter, interval, unit, season = cut_season(starttime))
) + facet_wrap(vars(season))
#> plotting with statistic mean and confidence interval of 0.95
# for faceting the variable must be included in the id_cols
gg_timevariation_wday(
data,
group = "parameter",
id_cols = grp(site, parameter, interval, unit, season = cut_season(starttime))
) + facet_wrap(vars(season))
#> plotting with statistic mean and confidence interval of 0.95
 # utility function to compose all plots together using patchwork
# for advanced use cases you should probably compose the plot yourself
# you can use ylim to start all y axis by 0. Lowering B speed up the process.
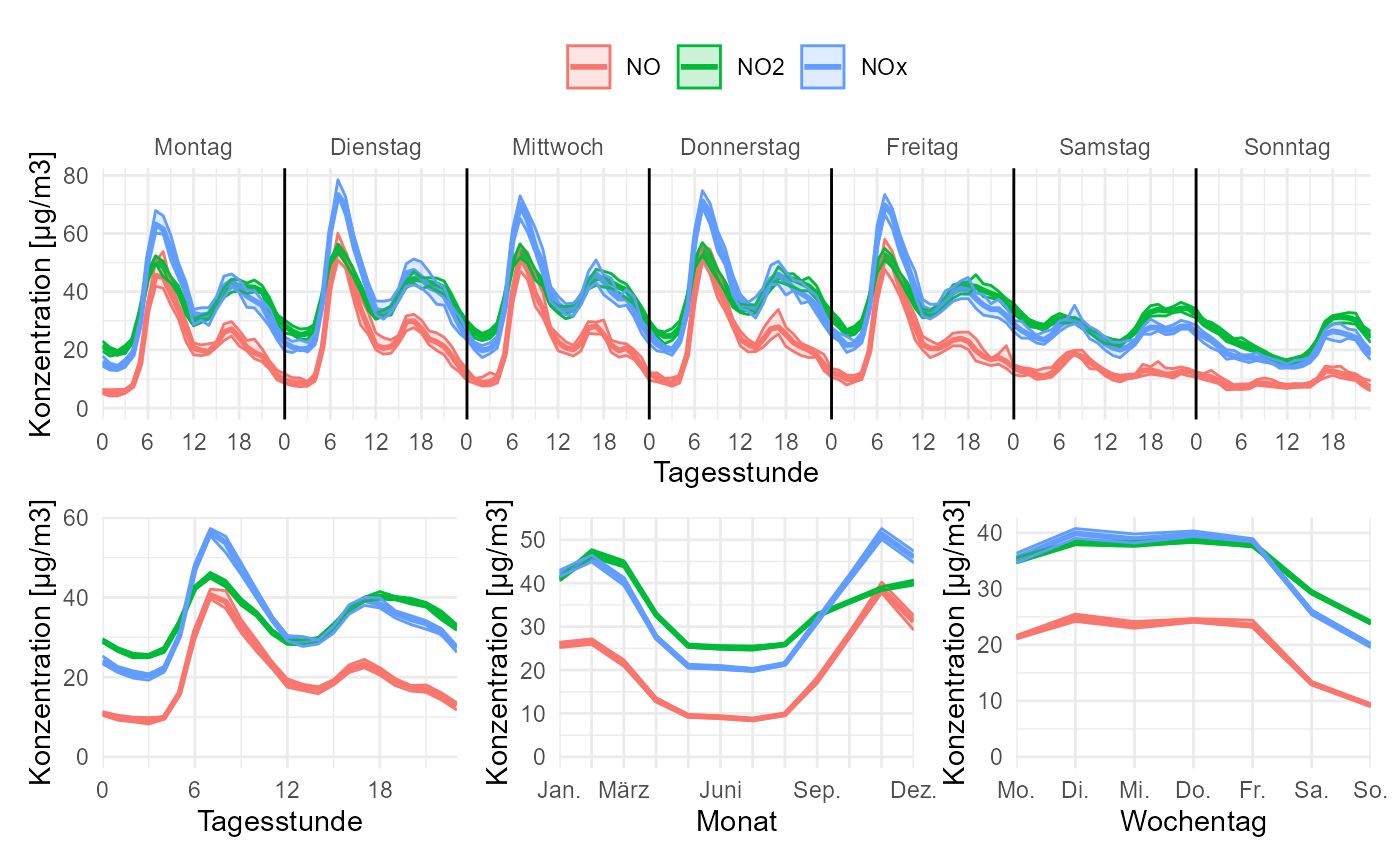
gg_timevariation(
data,
group = "parameter",
B = 10,
ylab = "Konzentration [µg/m3]",
ylim = c(0, NA)
)
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
# utility function to compose all plots together using patchwork
# for advanced use cases you should probably compose the plot yourself
# you can use ylim to start all y axis by 0. Lowering B speed up the process.
gg_timevariation(
data,
group = "parameter",
B = 10,
ylab = "Konzentration [µg/m3]",
ylim = c(0, NA)
)
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
#> plotting with statistic mean and confidence interval of 0.95
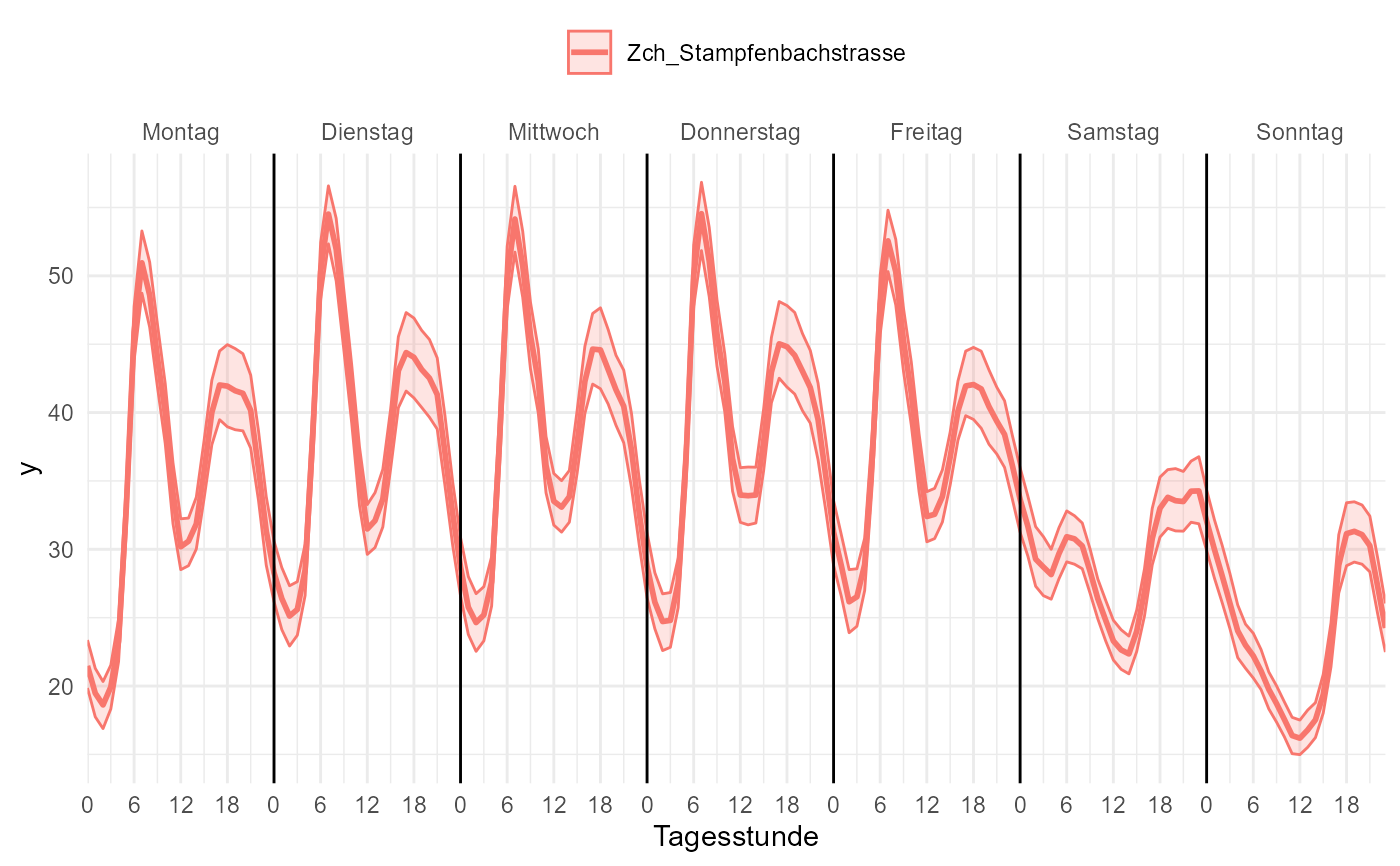
 # you can use wide data, but only with a single pollutant
data_wide <- rOstluft::rolf_to_openair(data)
gg_timevariation_wday_hour(
data_wide,
dt = date,
y = NO2,
group = "site",
id_cols = grp(site)
)
#> plotting with statistic mean and confidence interval of 0.95
# you can use wide data, but only with a single pollutant
data_wide <- rOstluft::rolf_to_openair(data)
gg_timevariation_wday_hour(
data_wide,
dt = date,
y = NO2,
group = "site",
id_cols = grp(site)
)
#> plotting with statistic mean and confidence interval of 0.95
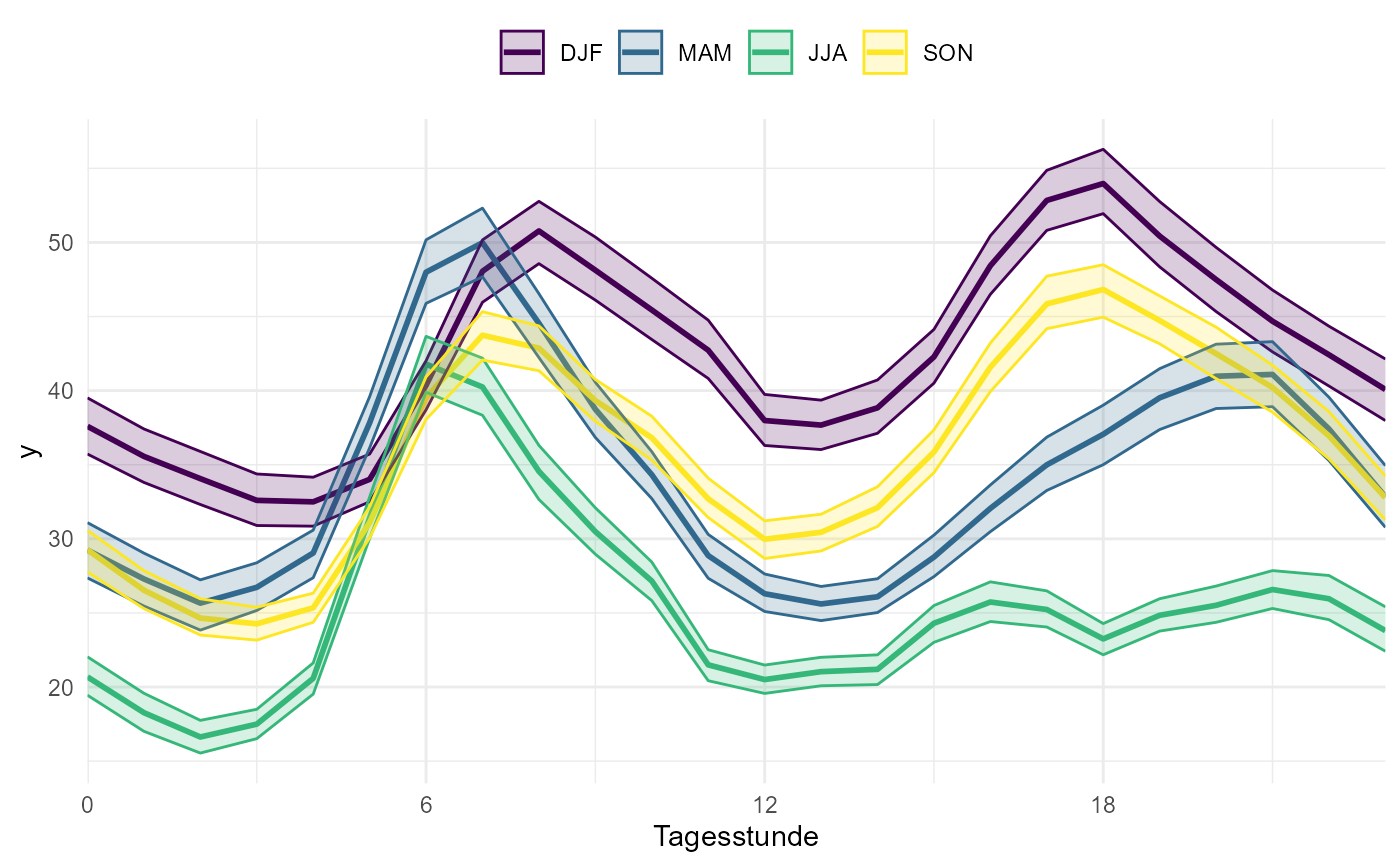
 # you can also use a function in id_cols to create groups
gg_timevariation_diurnal(
data_wide,
dt = date,
y = NO2,
group = "season",
id_cols = grp(site, season = cut_season(date))
)
#> plotting with statistic mean and confidence interval of 0.95
# you can also use a function in id_cols to create groups
gg_timevariation_diurnal(
data_wide,
dt = date,
y = NO2,
group = "season",
id_cols = grp(site, season = cut_season(date))
)
#> plotting with statistic mean and confidence interval of 0.95
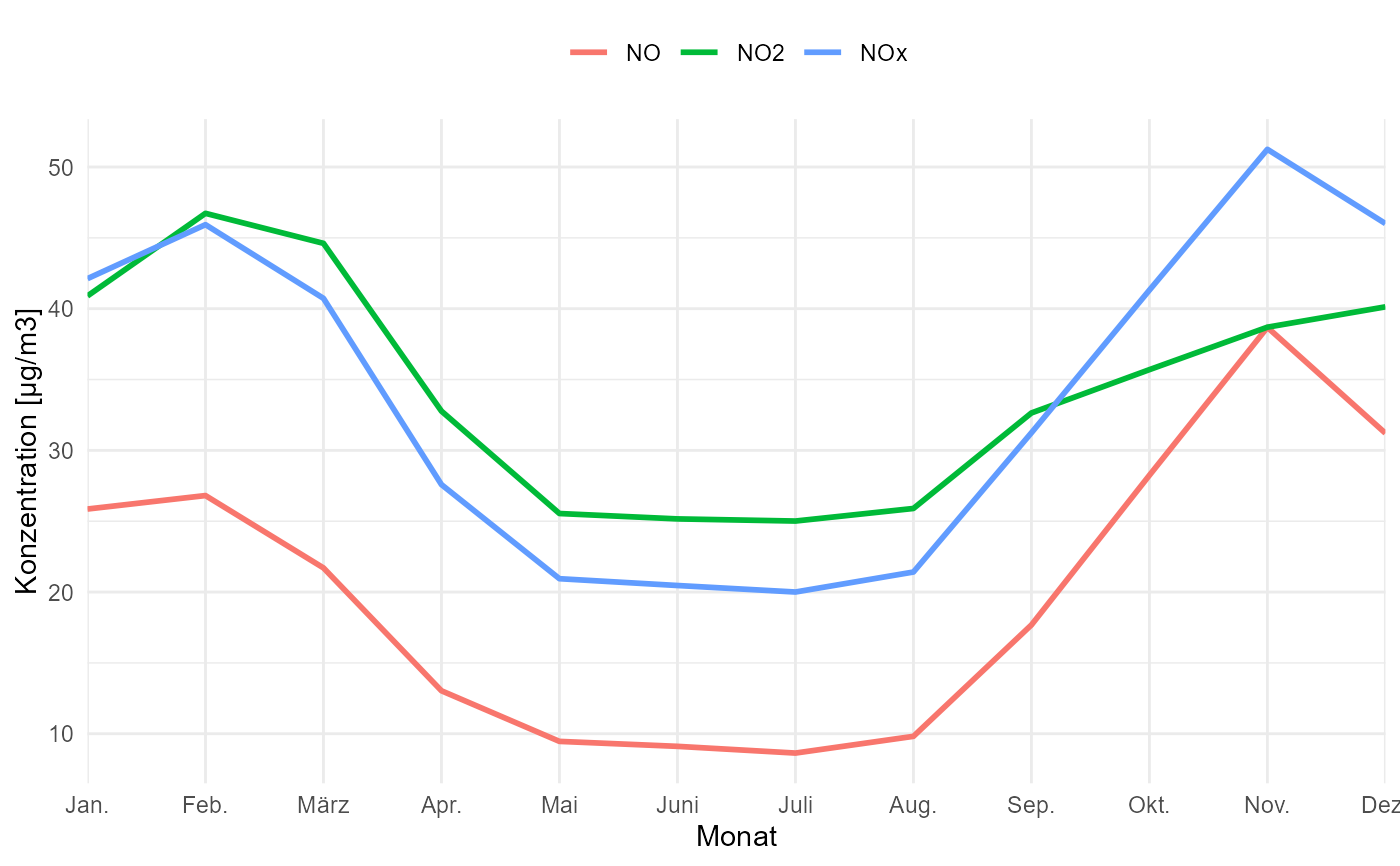
 # use an alternative geom
gg_timevariation_month(
data,
group = "parameter",
ylab = "Konzentration [µg/m3]",
B = 10,
geom_ci = geom_linerange(size = 2)
) +
scale_x_discrete(
expand = expansion(mult = 0.02)
)
#> plotting with statistic mean and confidence interval of 0.95
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
# use an alternative geom
gg_timevariation_month(
data,
group = "parameter",
ylab = "Konzentration [µg/m3]",
B = 10,
geom_ci = geom_linerange(size = 2)
) +
scale_x_discrete(
expand = expansion(mult = 0.02)
)
#> plotting with statistic mean and confidence interval of 0.95
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.